The PCA does an unsupervised dimensionality reduction, while the logistic regression does the prediction.
We use a GridSearchCV to set the dimensionality of the PCA
print(__doc__)
# Code source: Ga Varoquaux
# Modified for documentation by Jaques Grobler
# License: BSD 3 clause
import numpy as np
import matplotlib.pyplot as plt
from sklearn import linear_model, decomposition, datasets
from sklearn.pipeline import Pipeline
from sklearn.model_selection import GridSearchCV
logistic = linear_model.LogisticRegression()
pca = decomposition.PCA()
pipe = Pipeline(steps=[('pca', pca), ('logistic', logistic)])
digits = datasets.load_digits()
X_digits = digits.data
y_digits = digits.target
Plot the PCA spectrum
pca.fit(X_digits)
plt.figure(1, figsize=(4, 3))
plt.clf()
plt.axes([.2, .2, .7, .7])
plt.plot(pca.explained_variance_, linewidth=2)
plt.axis('tight')
plt.xlabel('n_components')
plt.ylabel('explained_variance_')
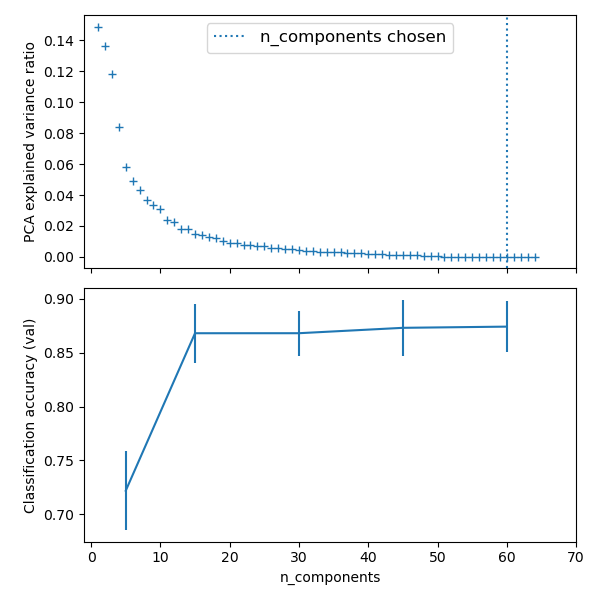
Prediction
n_components = [20, 40, 64]
Cs = np.logspace(-4, 4, 3)
#Parameters of pipelines can be set using ?__? separated parameter names:
estimator = GridSearchCV(pipe,
dict(pca__n_components=n_components,
logistic__C=Cs))
estimator.fit(X_digits, y_digits)
plt.axvline(estimator.best_estimator_.named_steps['pca'].n_components,
linestyle=':', label='n_components chosen')
plt.legend(prop=dict(size=12))
plt.show()

Total running time of the script: (0 minutes 9.258 seconds)
Download Python source code:
plot_digits_pipe.py
Download IPython notebook:
plot_digits_pipe.ipynb
Please login to continue.