-
class sklearn.linear_model.Perceptron(penalty=None, alpha=0.0001, fit_intercept=True, n_iter=5, shuffle=True, verbose=0, eta0=1.0, n_jobs=1, random_state=0, class_weight=None, warm_start=False)[source] -
Read more in the User Guide.
Parameters: penalty : None, ?l2? or ?l1? or ?elasticnet?
The penalty (aka regularization term) to be used. Defaults to None.
alpha : float
Constant that multiplies the regularization term if regularization is used. Defaults to 0.0001
fit_intercept : bool
Whether the intercept should be estimated or not. If False, the data is assumed to be already centered. Defaults to True.
n_iter : int, optional
The number of passes over the training data (aka epochs). Defaults to 5.
shuffle : bool, optional, default True
Whether or not the training data should be shuffled after each epoch.
random_state : int seed, RandomState instance, or None (default)
The seed of the pseudo random number generator to use when shuffling the data.
verbose : integer, optional
The verbosity level
n_jobs : integer, optional
The number of CPUs to use to do the OVA (One Versus All, for multi-class problems) computation. -1 means ?all CPUs?. Defaults to 1.
eta0 : double
Constant by which the updates are multiplied. Defaults to 1.
class_weight : dict, {class_label: weight} or ?balanced? or None, optional
Preset for the class_weight fit parameter.
Weights associated with classes. If not given, all classes are supposed to have weight one.
The ?balanced? mode uses the values of y to automatically adjust weights inversely proportional to class frequencies in the input data as
n_samples / (n_classes * np.bincount(y))warm_start : bool, optional
When set to True, reuse the solution of the previous call to fit as initialization, otherwise, just erase the previous solution.
Attributes: coef_ : array, shape = [1, n_features] if n_classes == 2 else [n_classes, n_features]
Weights assigned to the features.
intercept_ : array, shape = [1] if n_classes == 2 else [n_classes]
Constants in decision function.
See also
Notes
PerceptronandSGDClassifiershare the same underlying implementation. In fact,Perceptron()is equivalent toSGDClassifier(loss=?perceptron?, eta0=1, learning_rate=?constant?, penalty=None).References
https://en.wikipedia.org/wiki/Perceptron and references therein.
Methods
decision_function(X)Predict confidence scores for samples. densify()Convert coefficient matrix to dense array format. fit(X, y[, coef_init, intercept_init, ...])Fit linear model with Stochastic Gradient Descent. fit_transform(X[, y])Fit to data, then transform it. get_params([deep])Get parameters for this estimator. partial_fit(X, y[, classes, sample_weight])Fit linear model with Stochastic Gradient Descent. predict(X)Predict class labels for samples in X. score(X, y[, sample_weight])Returns the mean accuracy on the given test data and labels. set_params(\*args, \*\*kwargs)sparsify()Convert coefficient matrix to sparse format. transform(\*args, \*\*kwargs)DEPRECATED: Support to use estimators as feature selectors will be removed in version 0.19. -
__init__(penalty=None, alpha=0.0001, fit_intercept=True, n_iter=5, shuffle=True, verbose=0, eta0=1.0, n_jobs=1, random_state=0, class_weight=None, warm_start=False)[source]
-
decision_function(X)[source] -
Predict confidence scores for samples.
The confidence score for a sample is the signed distance of that sample to the hyperplane.
Parameters: X : {array-like, sparse matrix}, shape = (n_samples, n_features)
Samples.
Returns: array, shape=(n_samples,) if n_classes == 2 else (n_samples, n_classes) :
Confidence scores per (sample, class) combination. In the binary case, confidence score for self.classes_[1] where >0 means this class would be predicted.
-
densify()[source] -
Convert coefficient matrix to dense array format.
Converts the
coef_member (back) to a numpy.ndarray. This is the default format ofcoef_and is required for fitting, so calling this method is only required on models that have previously been sparsified; otherwise, it is a no-op.Returns: self: estimator :
-
fit(X, y, coef_init=None, intercept_init=None, sample_weight=None)[source] -
Fit linear model with Stochastic Gradient Descent.
Parameters: X : {array-like, sparse matrix}, shape (n_samples, n_features)
Training data
y : numpy array, shape (n_samples,)
Target values
coef_init : array, shape (n_classes, n_features)
The initial coefficients to warm-start the optimization.
intercept_init : array, shape (n_classes,)
The initial intercept to warm-start the optimization.
sample_weight : array-like, shape (n_samples,), optional
Weights applied to individual samples. If not provided, uniform weights are assumed. These weights will be multiplied with class_weight (passed through the constructor) if class_weight is specified
Returns: self : returns an instance of self.
-
fit_transform(X, y=None, **fit_params)[source] -
Fit to data, then transform it.
Fits transformer to X and y with optional parameters fit_params and returns a transformed version of X.
Parameters: X : numpy array of shape [n_samples, n_features]
Training set.
y : numpy array of shape [n_samples]
Target values.
Returns: X_new : numpy array of shape [n_samples, n_features_new]
Transformed array.
-
get_params(deep=True)[source] -
Get parameters for this estimator.
Parameters: deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: params : mapping of string to any
Parameter names mapped to their values.
-
partial_fit(X, y, classes=None, sample_weight=None)[source] -
Fit linear model with Stochastic Gradient Descent.
Parameters: X : {array-like, sparse matrix}, shape (n_samples, n_features)
Subset of the training data
y : numpy array, shape (n_samples,)
Subset of the target values
classes : array, shape (n_classes,)
Classes across all calls to partial_fit. Can be obtained by via
np.unique(y_all), where y_all is the target vector of the entire dataset. This argument is required for the first call to partial_fit and can be omitted in the subsequent calls. Note that y doesn?t need to contain all labels inclasses.sample_weight : array-like, shape (n_samples,), optional
Weights applied to individual samples. If not provided, uniform weights are assumed.
Returns: self : returns an instance of self.
-
predict(X)[source] -
Predict class labels for samples in X.
Parameters: X : {array-like, sparse matrix}, shape = [n_samples, n_features]
Samples.
Returns: C : array, shape = [n_samples]
Predicted class label per sample.
-
score(X, y, sample_weight=None)[source] -
Returns the mean accuracy on the given test data and labels.
In multi-label classification, this is the subset accuracy which is a harsh metric since you require for each sample that each label set be correctly predicted.
Parameters: X : array-like, shape = (n_samples, n_features)
Test samples.
y : array-like, shape = (n_samples) or (n_samples, n_outputs)
True labels for X.
sample_weight : array-like, shape = [n_samples], optional
Sample weights.
Returns: score : float
Mean accuracy of self.predict(X) wrt. y.
-
sparsify()[source] -
Convert coefficient matrix to sparse format.
Converts the
coef_member to a scipy.sparse matrix, which for L1-regularized models can be much more memory- and storage-efficient than the usual numpy.ndarray representation.The
intercept_member is not converted.Returns: self: estimator : Notes
For non-sparse models, i.e. when there are not many zeros in
coef_, this may actually increase memory usage, so use this method with care. A rule of thumb is that the number of zero elements, which can be computed with(coef_ == 0).sum(), must be more than 50% for this to provide significant benefits.After calling this method, further fitting with the partial_fit method (if any) will not work until you call densify.
-
transform(*args, **kwargs)[source] -
DEPRECATED: Support to use estimators as feature selectors will be removed in version 0.19. Use SelectFromModel instead.
Reduce X to its most important features.
Usescoef_orfeature_importances_to determine the most important features. For models with acoef_for each class, the absolute sum over the classes is used.Parameters: X : array or scipy sparse matrix of shape [n_samples, n_features]
The input samples.
- threshold
-
The threshold value to use for feature selection. Features whose importance is greater or equal are kept while the others are discarded. If ?median? (resp. ?mean?), then the threshold value is the median (resp. the mean) of the feature importances. A scaling factor (e.g., ?1.25*mean?) may also be used. If None and if available, the object attribute
thresholdis used. Otherwise, ?mean? is used by default.
Returns:
X_r : array of shape [n_samples, n_selected_features]
The input samples with only the selected features.
-
linear_model.Perceptron()
Examples using
2025-01-10 15:47:30
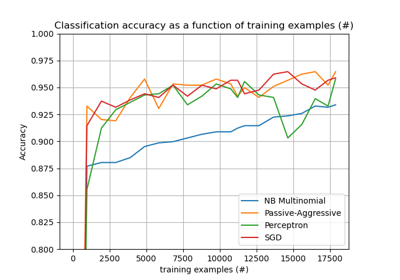
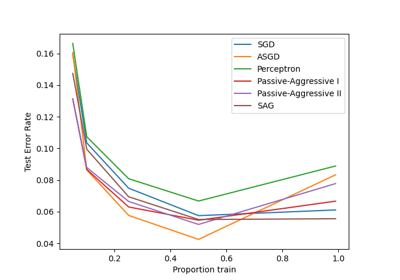

Please login to continue.